DIGITAL NARRATIVES BEYOND HUMAN SCALE
Every fourth breath we take is sponsored by diatoms since these photosynthetic unicellular organisms produce about one-fourth of the world's oxygen. Although these microscopic organisms are so incredibly important for our carbon cycle, few people apart from biologists and microscopists are familiar with them. The reason for their general unfamiliarity is probably their size. Diatoms are between 1 and 500 micrometers in size and only become visible under the microscope. In the past, however, not only biologists but also artists have become enthusiastic about the diverse geometric forms of the glassy unicellular organisms. This work shows an approach to generatively create digital diatom models based on real diatoms that are identified live under a light microscope using deep-learning-based image detection. All digital diatoms are generated and rendered in real-time in 4k.
Every fourth breath we take is sponsored by diatoms since these photosynthetic unicellular organisms produce about one-fourth of the world's oxygen. Although these microscopic organisms are so incredibly important for our carbon cycle, few people apart from biologists and microscopists are familiar with them. The reason for their general unfamiliarity is probably their size. Diatoms are between 1 and 500 micrometers in size and only become visible under the microscope. In the past, however, not only biologists but also artists have become enthusiastic about the diverse geometric forms of the glassy unicellular organisms. This work shows an approach to generatively create digital diatom models based on real diatoms that are identified live under a light microscope using deep-learning-based image detection. All digital diatoms are generated and rendered in real-time in 4k.


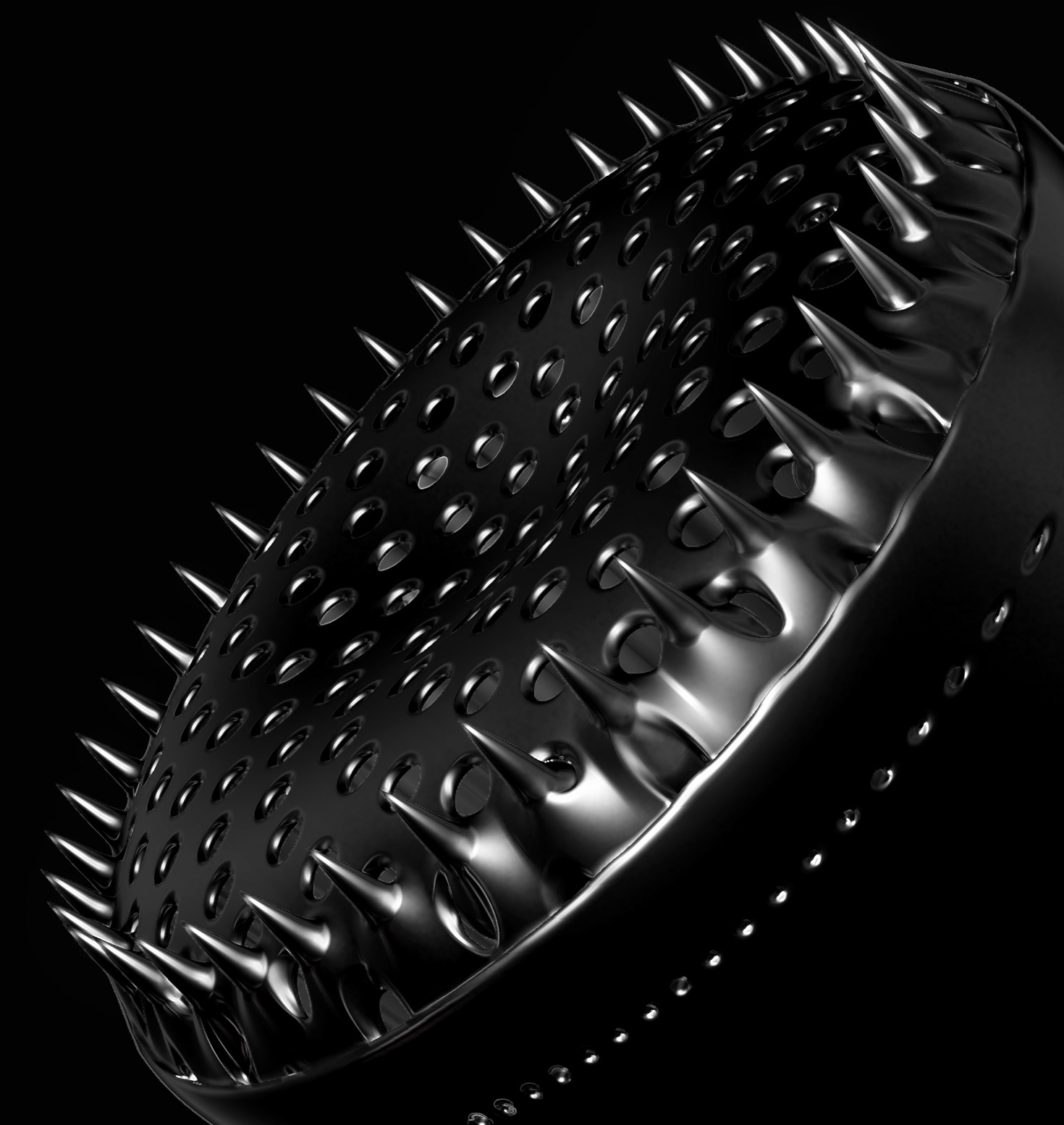
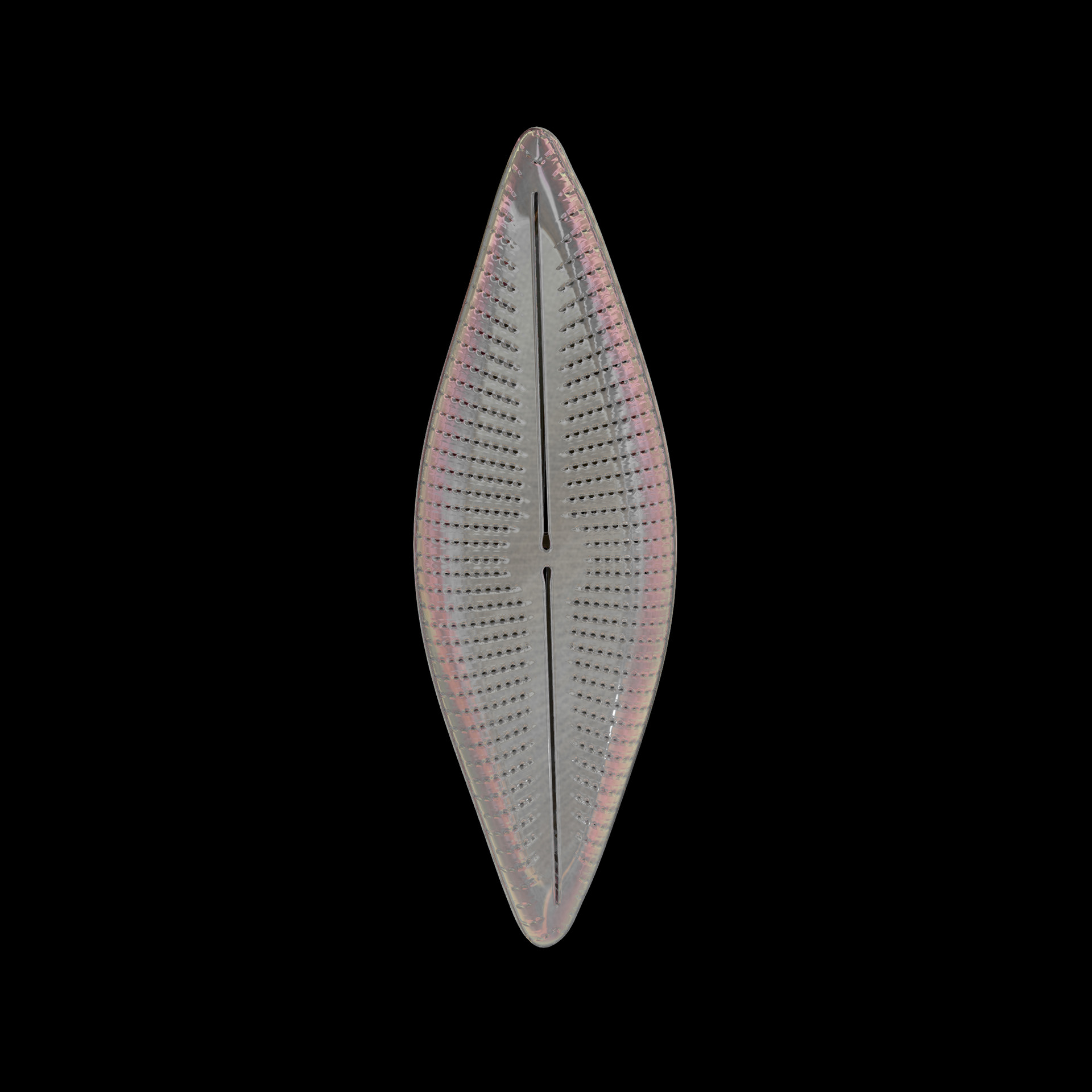
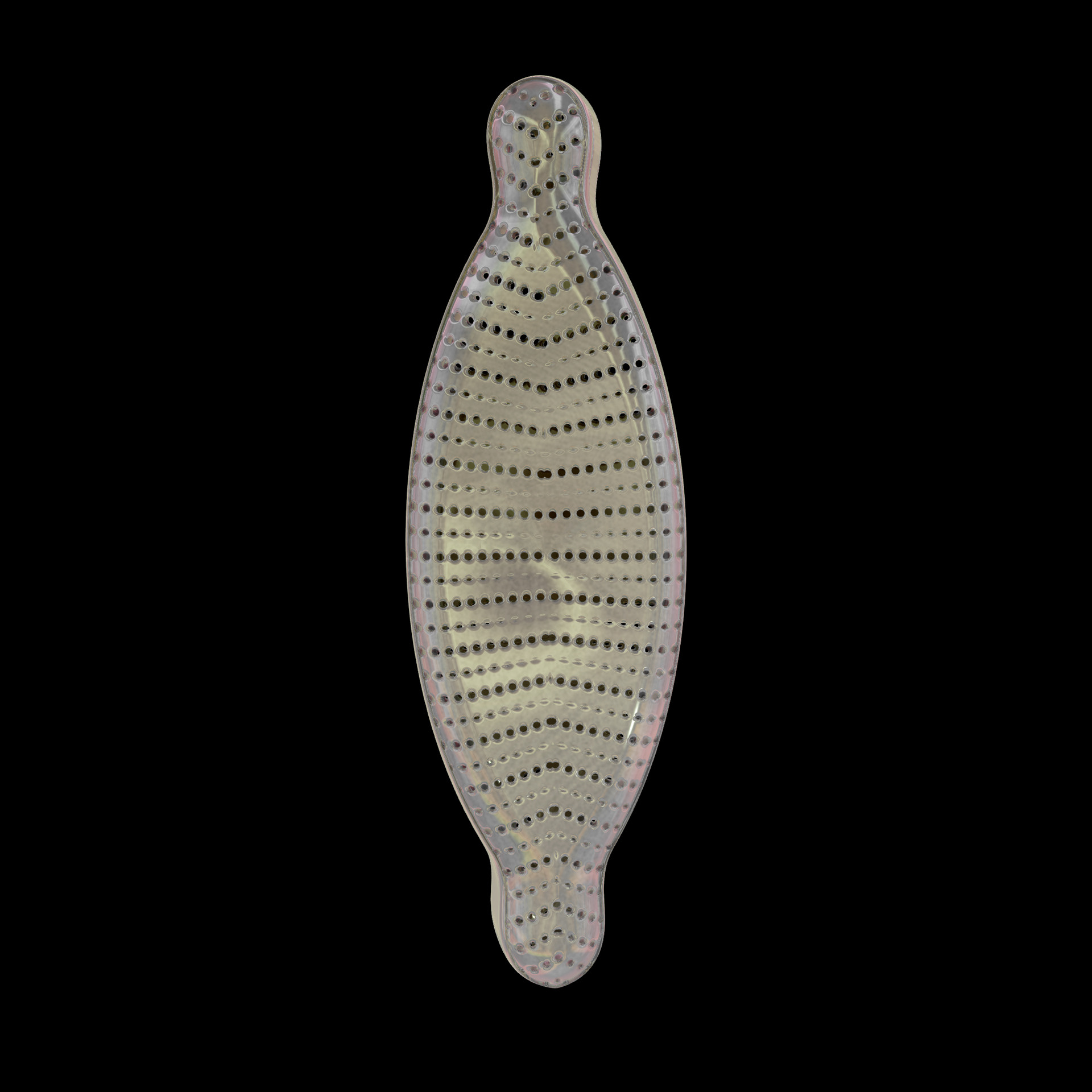
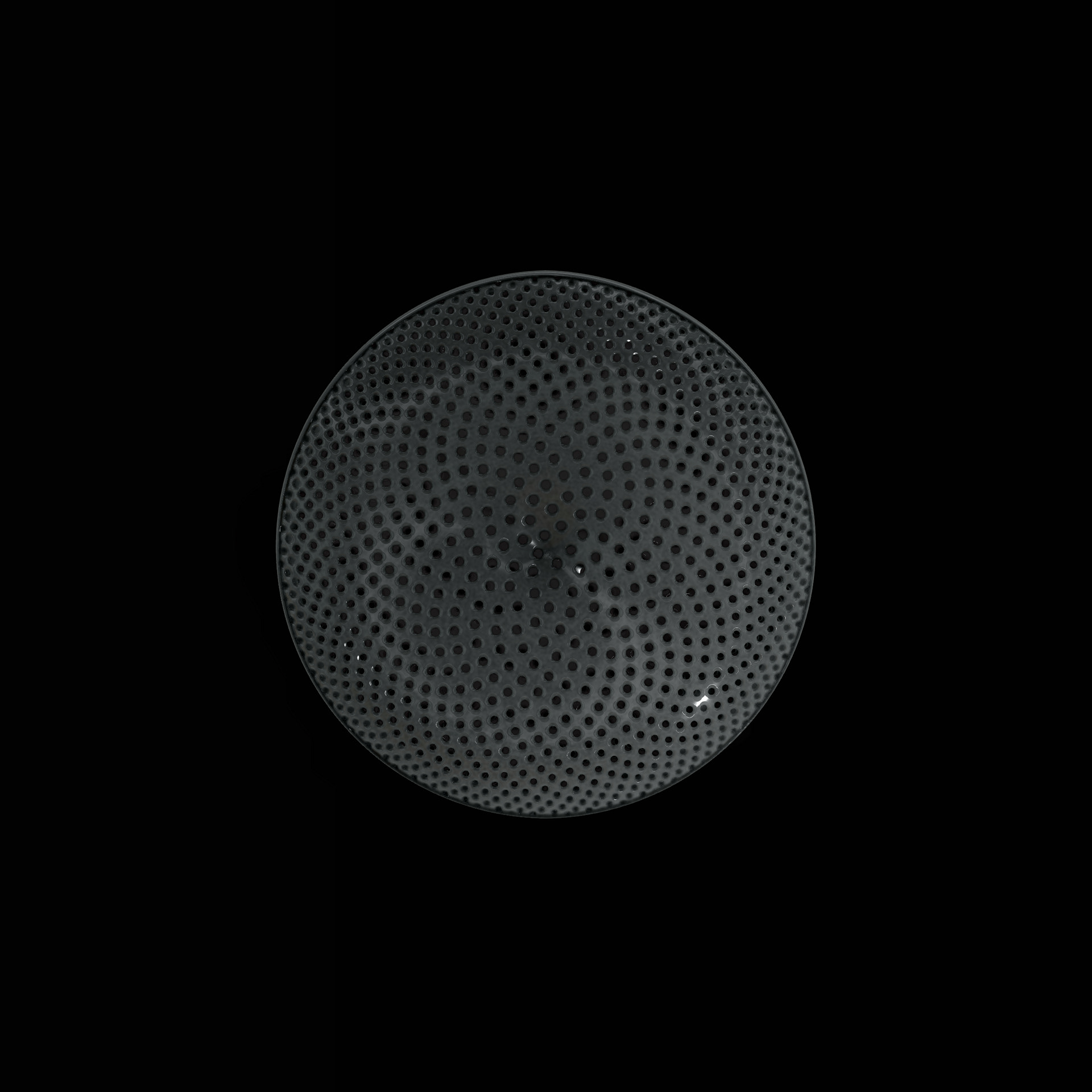
Diatoms show an extraordinary diversity of forms and species. They are basically divided into two large groups: Centric diatoms with radial symmetry and pennate diatoms with an elongated shape and bilateral symmetry. The generative diatom models try to represent this diversity of shapes algorithmically.
The frustules, i.e. the shells, of diatoms consist of silicon dioxide, the same basic material that industrially manufactured drinking or window glasses are made of, but also naturally occurring opals. The materiality of diatom frustules can be thought of as a mixture of the transparency of glass and the iridescence of opals. Since the complex transparency and refraction of light of the frustules could not be visualised in real time, the materiality of the digital diatoms mainly tried to represent the iridescent ability of the frustules, similar to opals. Because of this ability, diatoms are often referred to as "Living Opals" or "Jewels of the Sea". The viewer also has the option of looking at the frustules in a high-contrast black-and-white representation to view the surface structure even better.
Colony formation
Some diatom species form colonies that also adopt a species-specific formation. Some centric diatoms, so-called filamentous diatoms, hang together in long chains.
"What a contrast! In the 19th century, diatoms were arranged into ornamental patterns under the microscope. In the 21st century, AI recognizes diatoms and algorithms allow it to digitally and interactively play with them. But the infinite variety of diatom forms has fascinated people throughout the ages."
(Matthias Burba about this work)
(Matthias Burba about this work)
Diatoms are found in any body of water, whether salt or fresh water, flowing or stagnant, they can even be found in a puddle or soil moisture. The following videos show microscopic footage of some of the collected live samples under the light microscope, both in darkfield and brightfield.

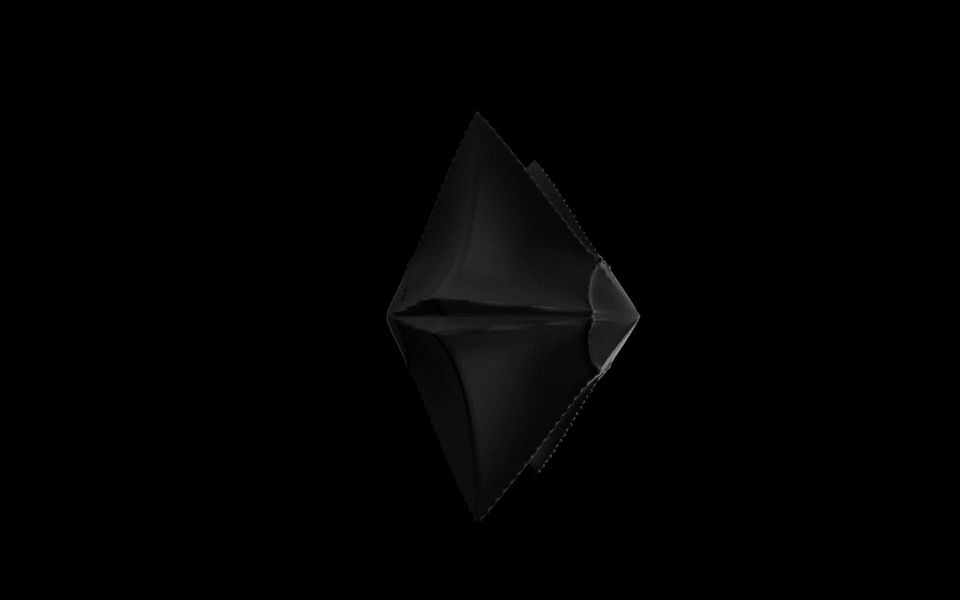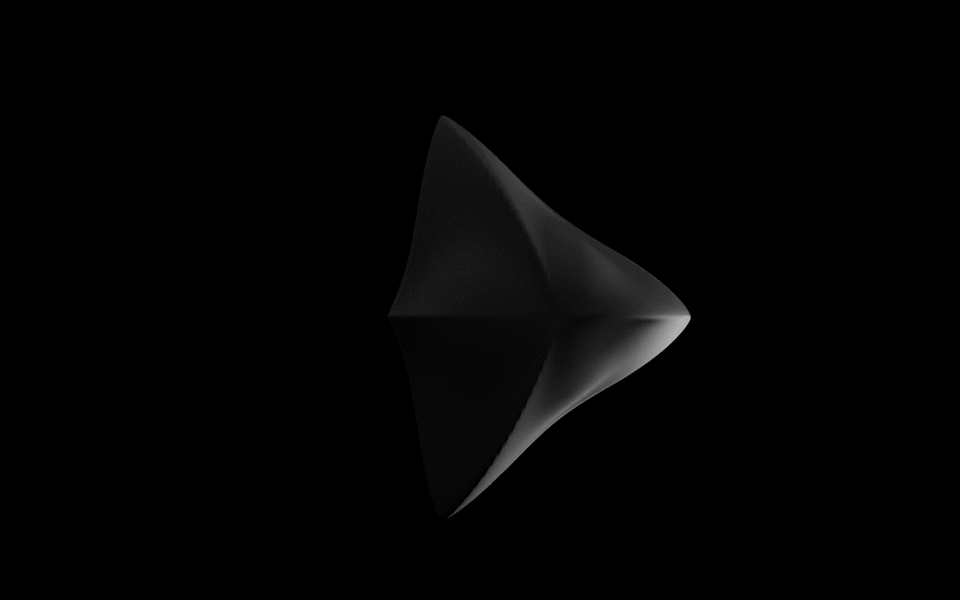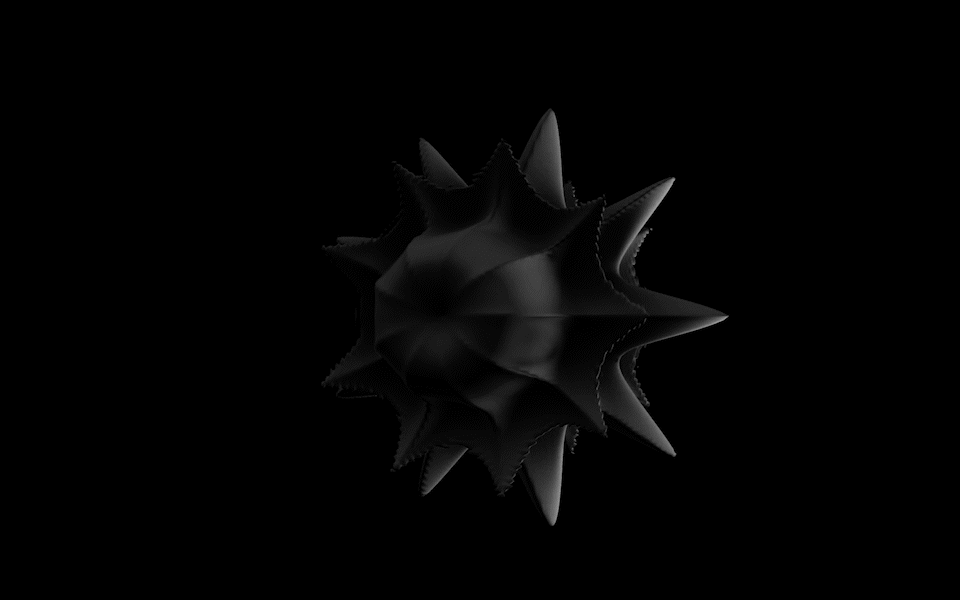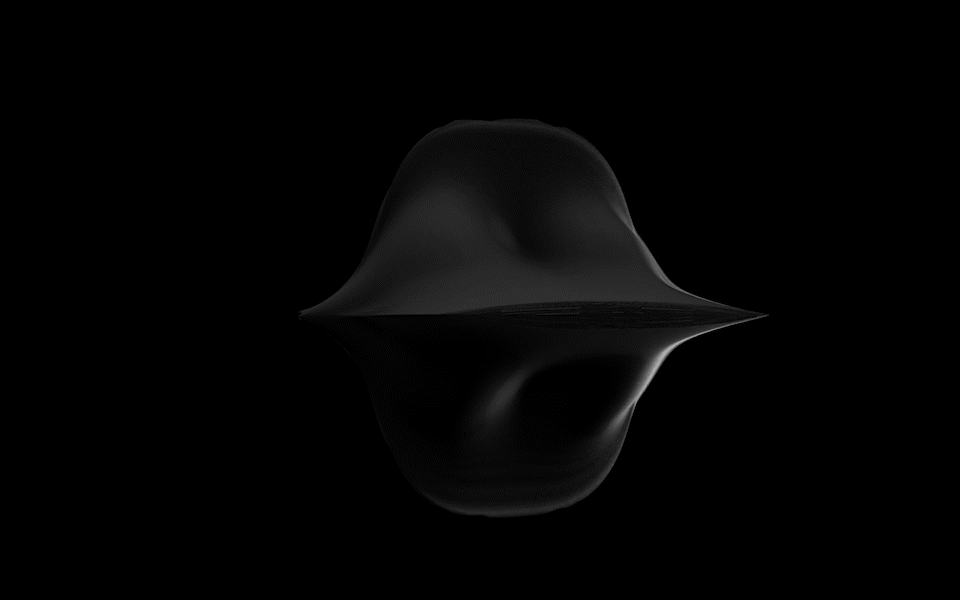
The Super Formula
Johann Gielis (2003) has set up a geometric formula, which makes it possible to produce an incredible variety of natural shapes with only a handful of parameters. As Gielis notes in later publications, in addition to shells, flowers, or leaves, typical diatom frustules can also be modeled with the formula, both in 2D and 3D.
For the calculation of the spatial form of digital diatoms, Gieli's so-called super formula was therefore used in this project. The superformula was implemented in a shader to make it run smoothly on the GPU. All parameters can be influenced dynamically and in real-time.
For the calculation of the spatial form of digital diatoms, Gieli's so-called super formula was therefore used in this project. The superformula was implemented in a shader to make it run smoothly on the GPU. All parameters can be influenced dynamically and in real-time.
Surface Structure
The fine pore structure, as well as the appearance of species-specific features such as spica, raphe, or striae, usually make it possible to distinguish between different species under the light microscope. Nevertheless, these differences between different diatom species are extremely small and the structure and microstructure of diatom frustules are extremely detailed. In order to generatively model the different features of the pore structure, two different algorithms were used. Firstly, classical circle packing, inspired by Frei Otto's approach of representing the structure of diatoms on the closest packing of pneus or simplified spheres, was used. Secondly, in order to be able to represent some diatom species, the point distribution according to Vogel (1979) was also used, which is based on the distribution of the golden ratio.





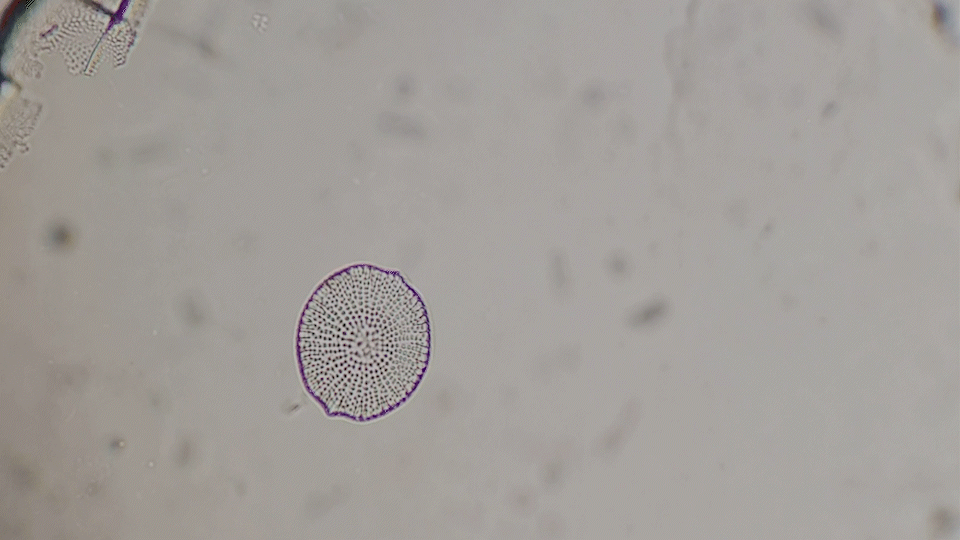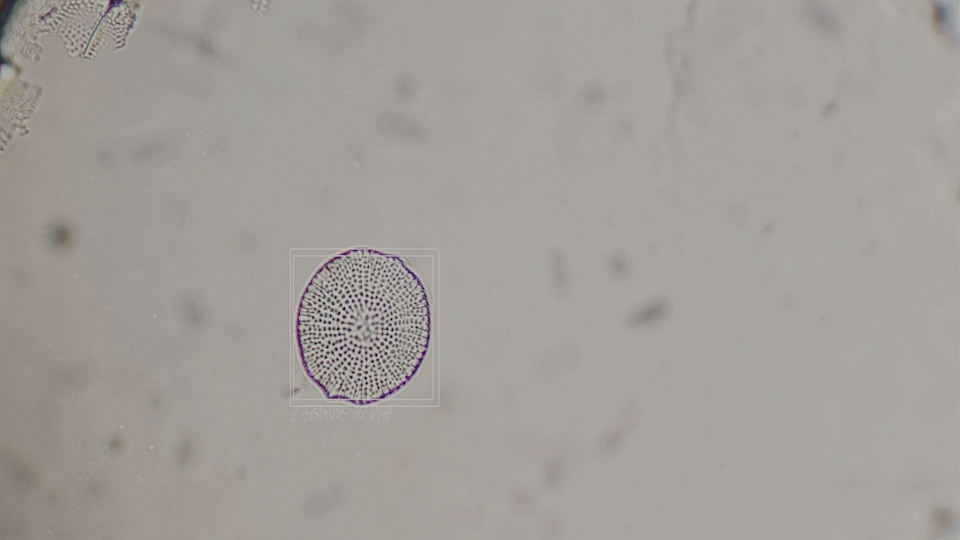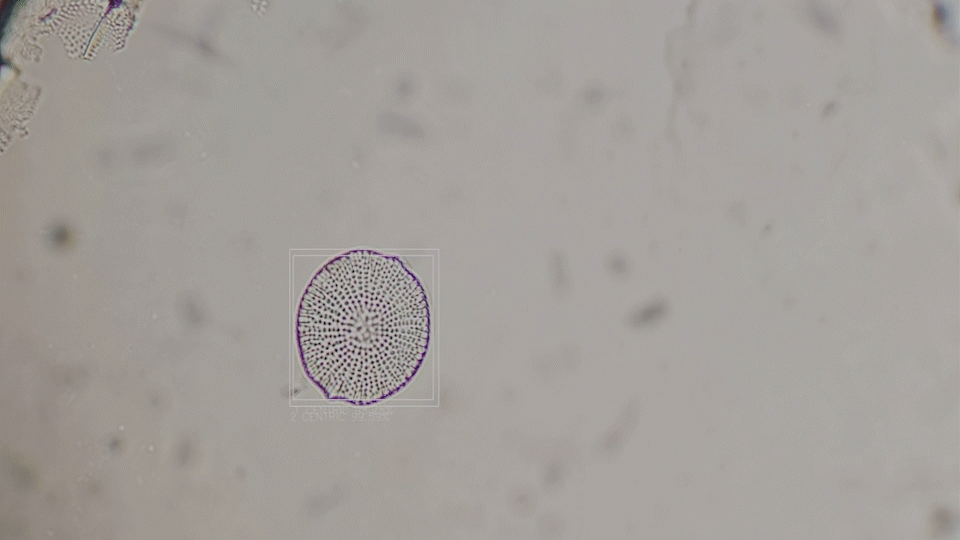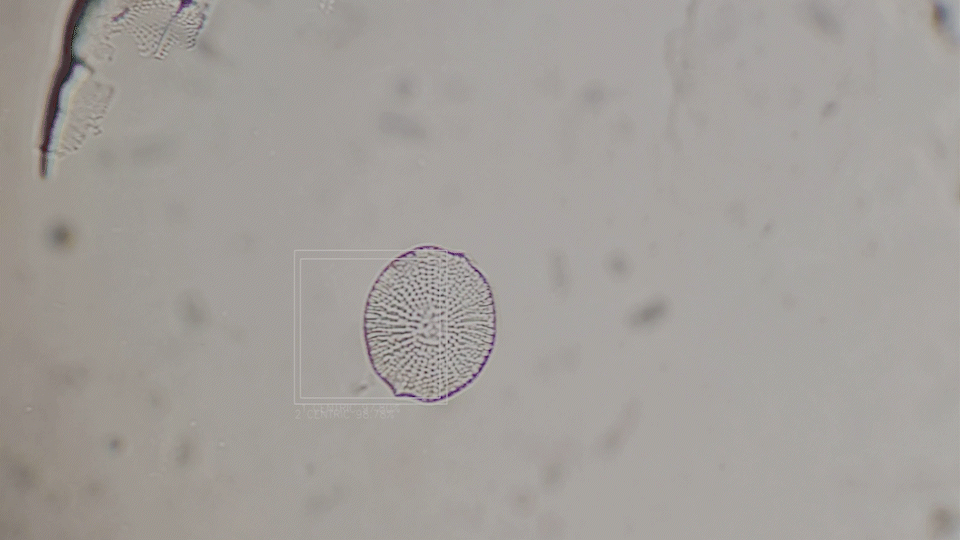
Deep-Learning based diatom detection
The generative diatoms in this project were created based on real diatoms visible under a microscope. For this purpose, a deep-learning based image detection model was trained to recognise different diatoms under a light microscope in real time. Since the differentiation into all different diatom species (current estimates are around 100,000 different species) is impossible within the scope of this work, the YOLO image detection model was only trained to recognise diatoms within a microscopic sample and to assign them to one of the higher-level taxonomic groups of diatoms. The image dataset for training the image detection model comprised approximately 6000 microscopic images of diatoms, both from live samples and from specimens taken specifically for this project. A Zeiss Standard 14 light microscope with an adapted smartphone camera and a self-built Rasperry Pi camera were used.




Installation
The resulting installation consists of a microscope table in which four monitors are embedded. Both the live image of the microscope and the generative diatoms are visible on the screens. The viewers can choose from different prepared diatom slides themselves or freshly pipette from water samples. The diatom samples can be moved directly by the microscope's mechanics, while the digital diatoms can be rotated and viewed from all sides via hand tracking using a leap motion.
The resulting installation consists of a microscope table in which four monitors are embedded. Both the live image of the microscope and the generative diatoms are visible on the screens. The viewers can choose from different prepared diatom slides themselves or freshly pipette from water samples. The diatom samples can be moved directly by the microscope's mechanics, while the digital diatoms can be rotated and viewed from all sides via hand tracking using a leap motion.



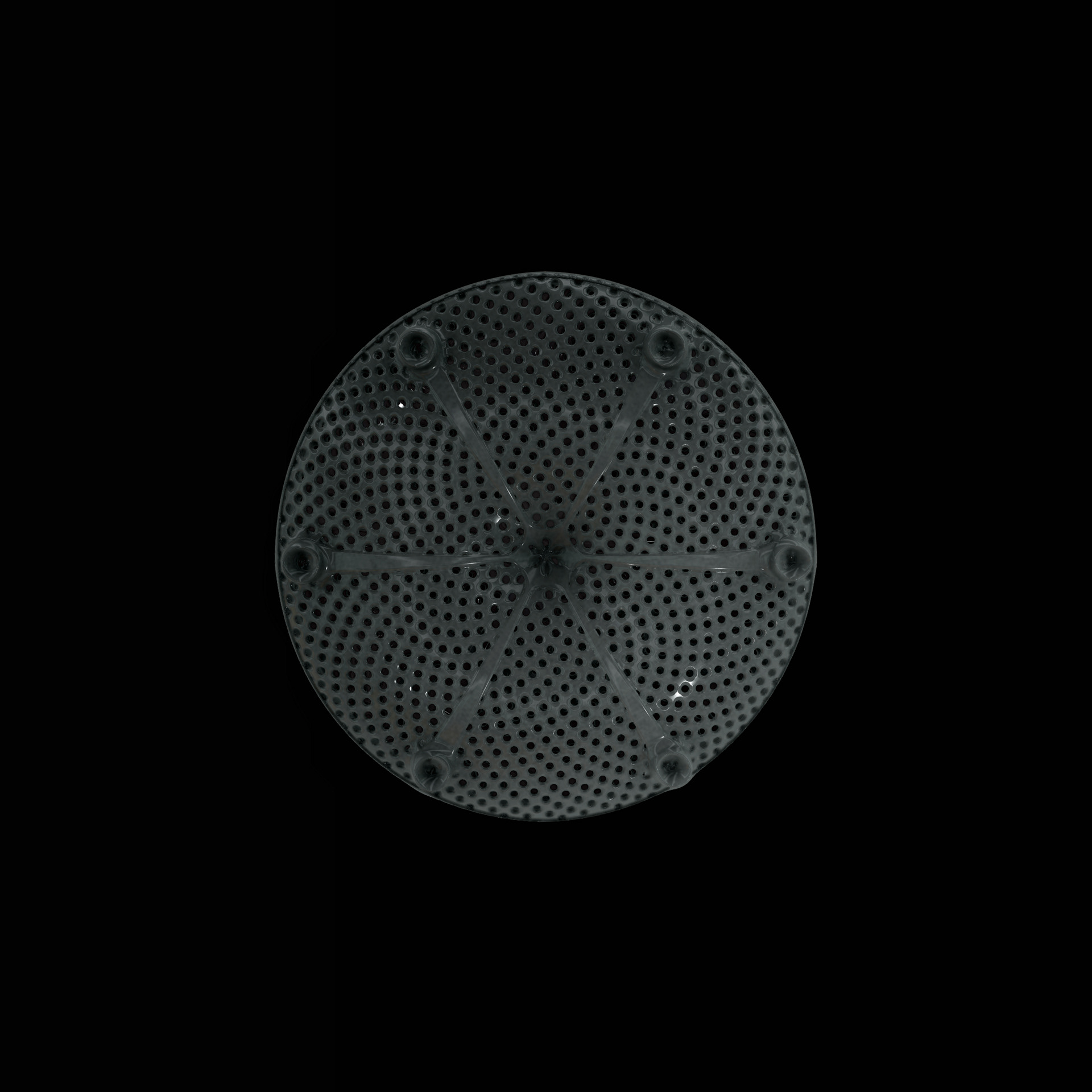
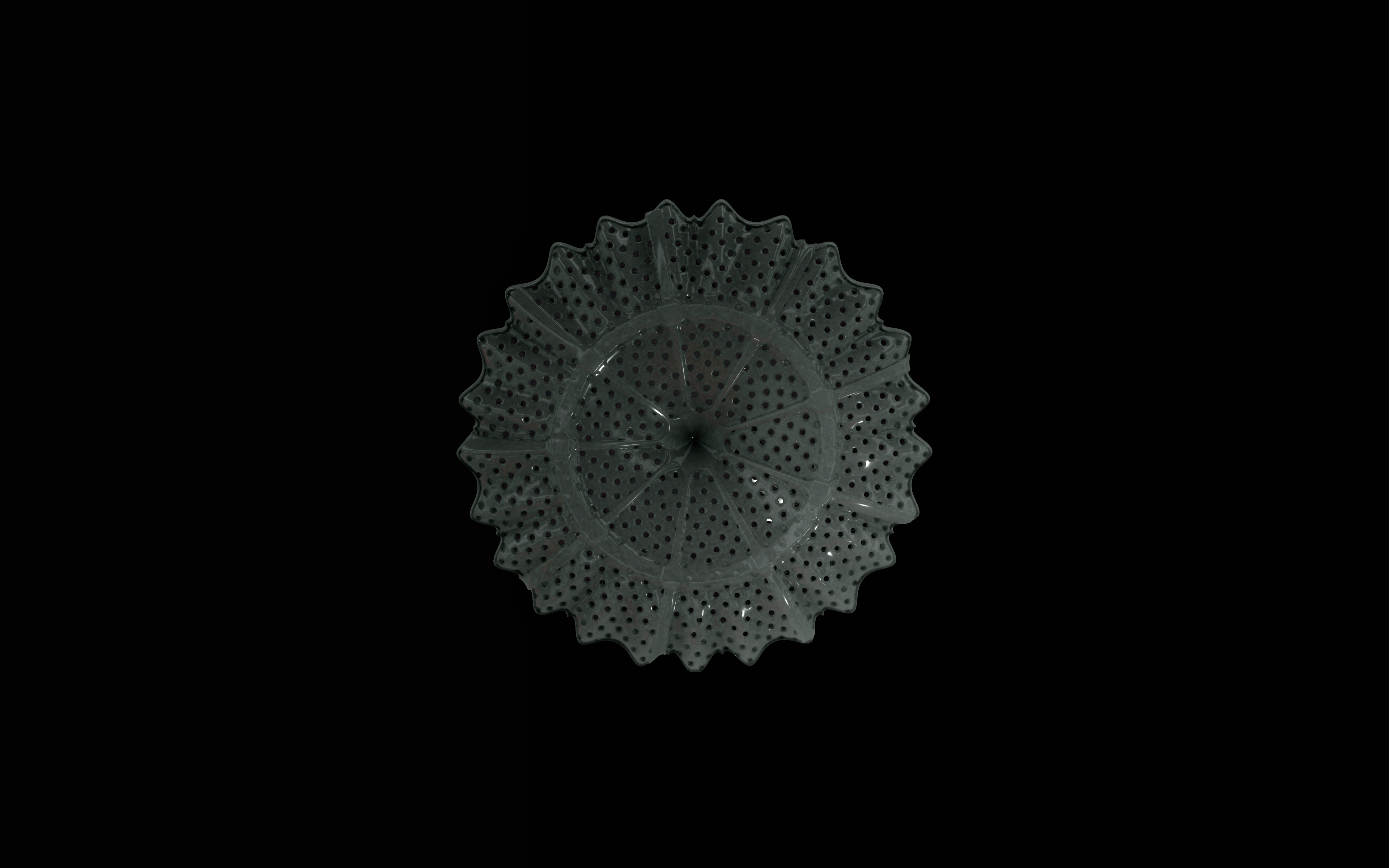
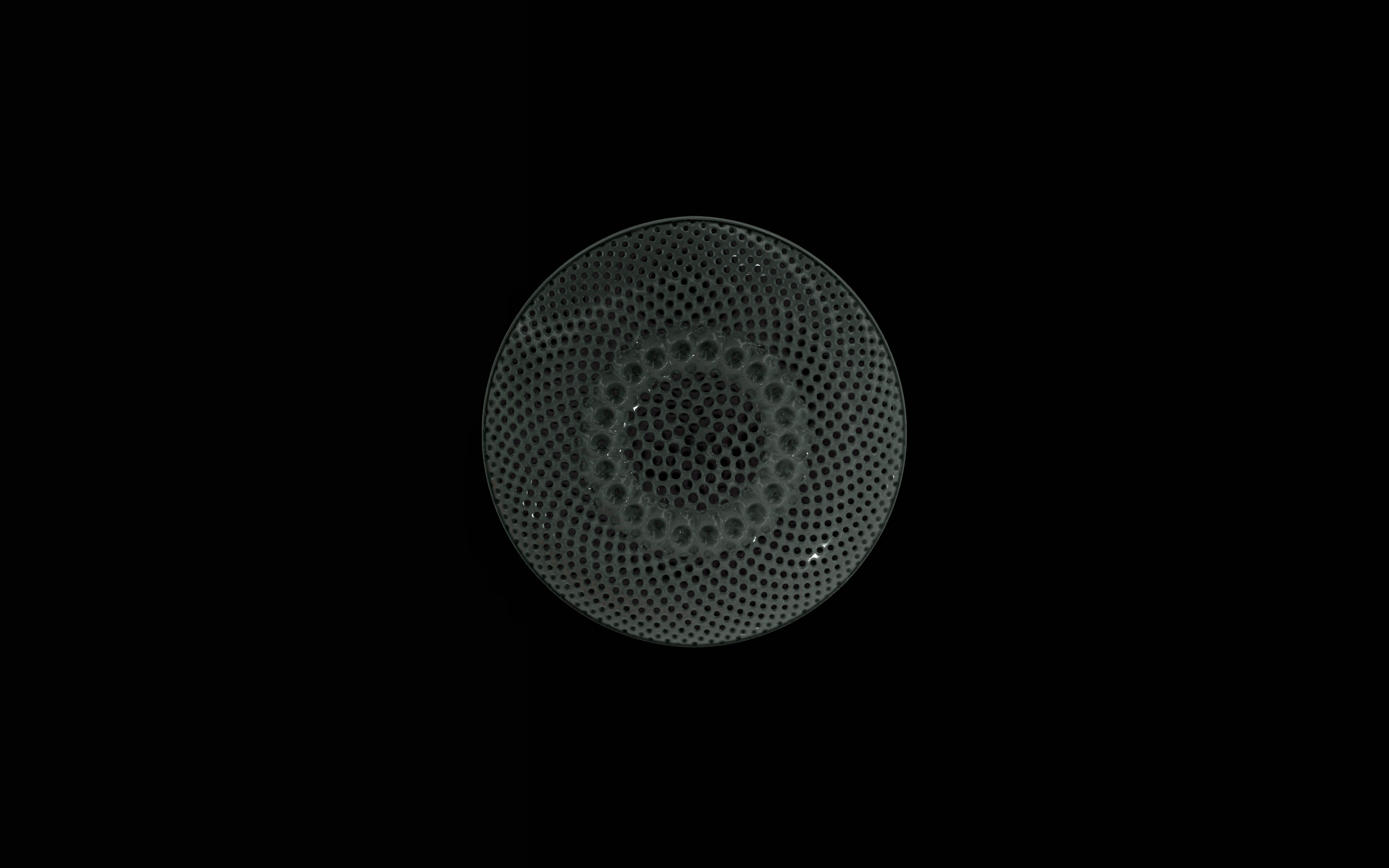
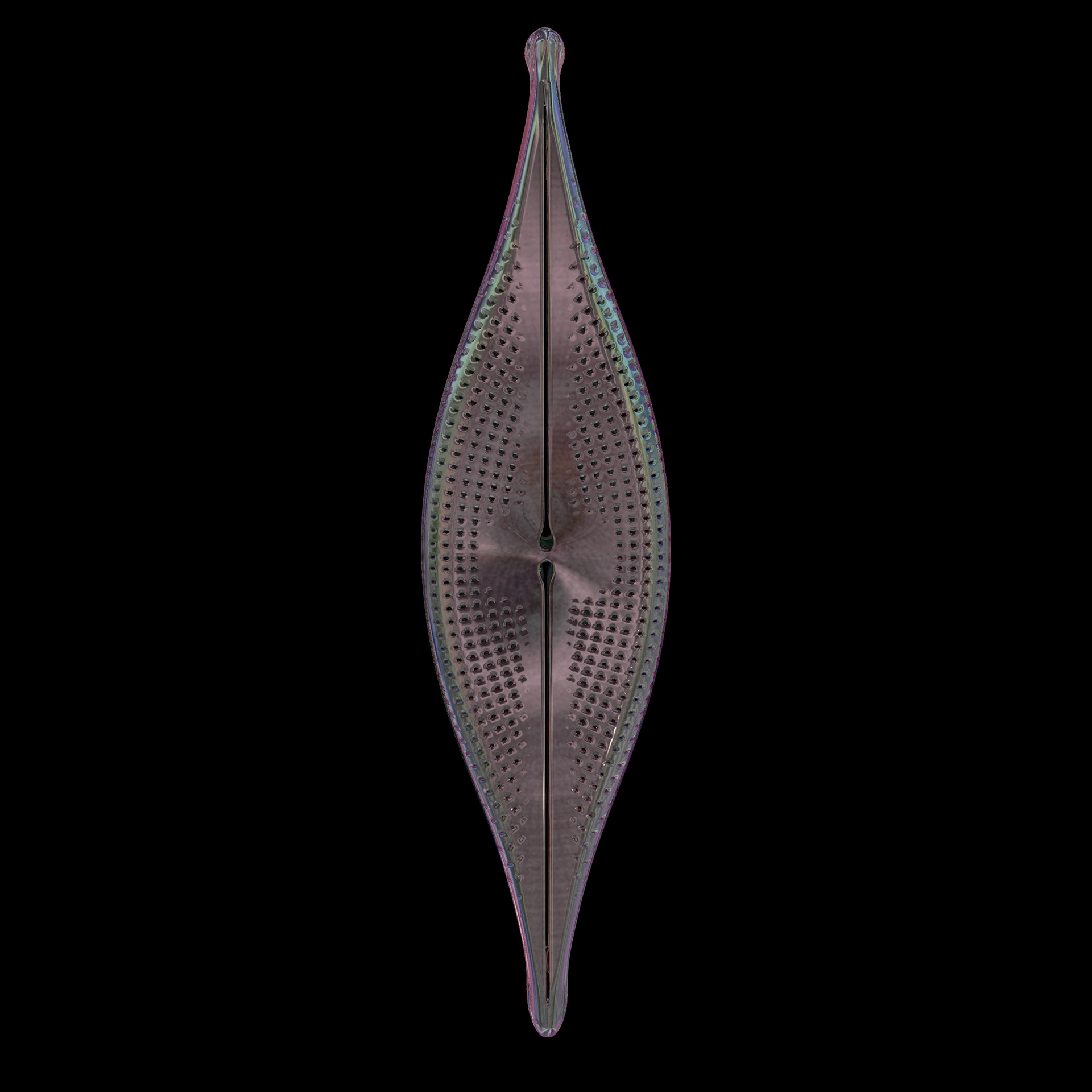

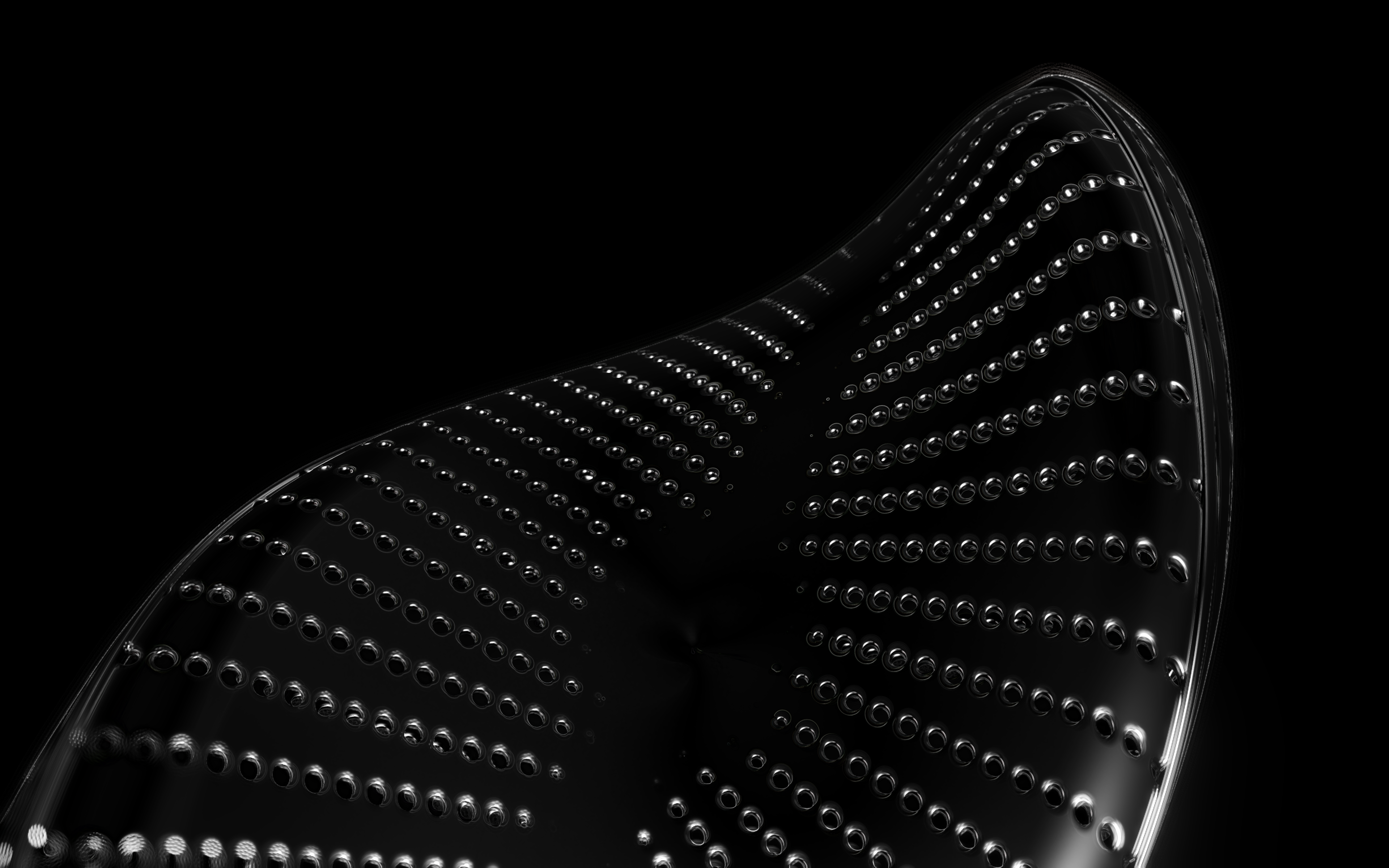

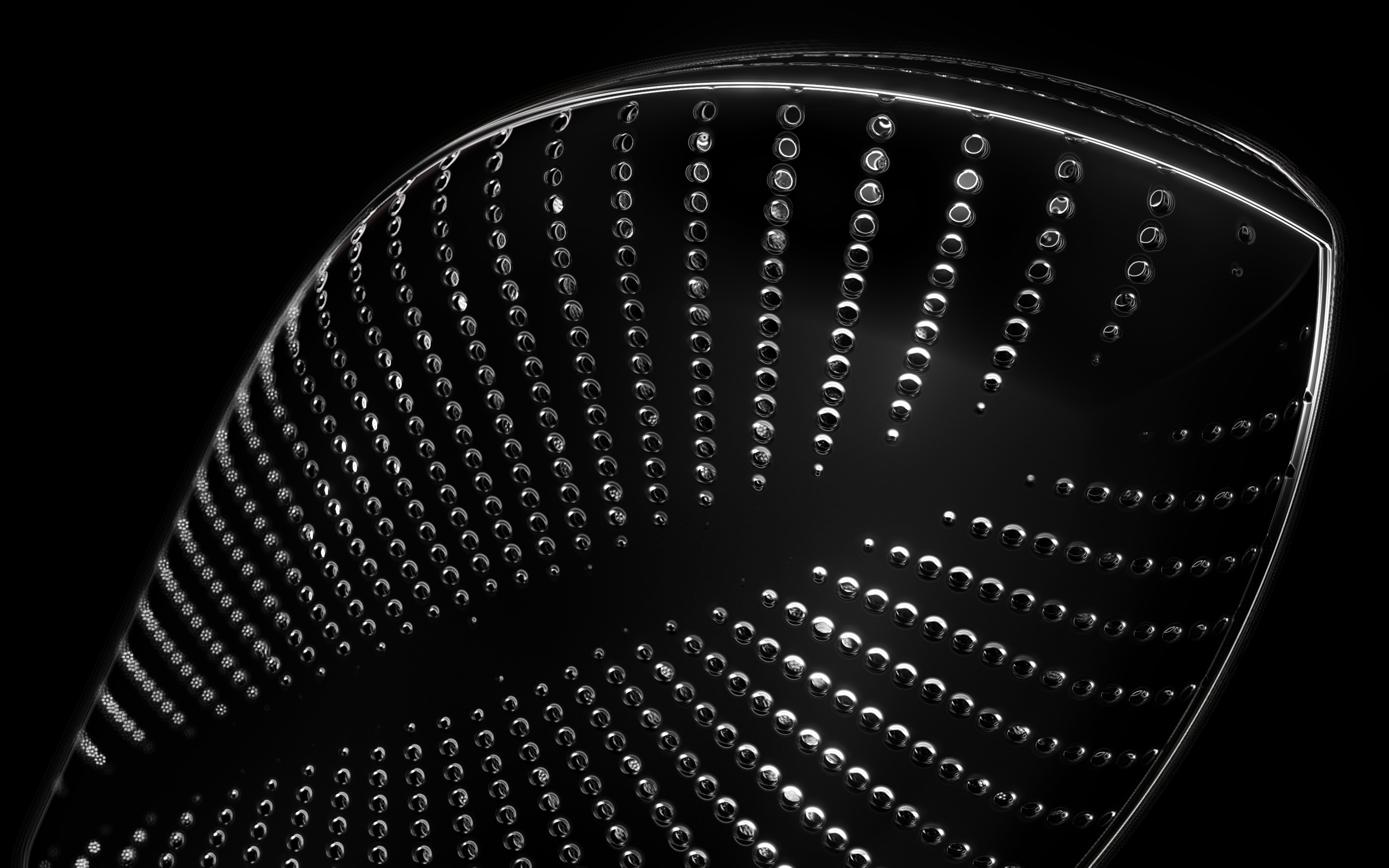


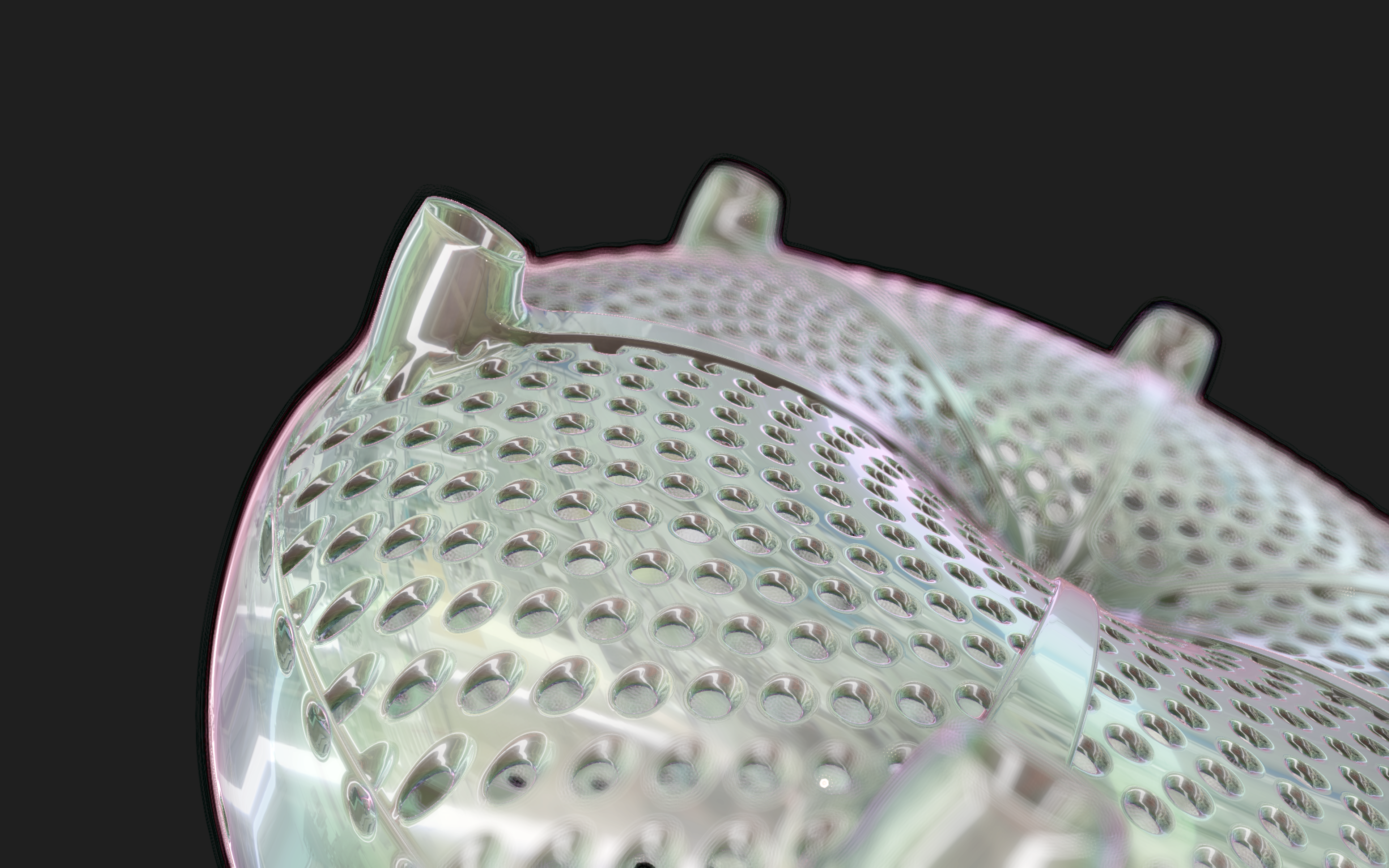




Jewels of the Sea was awarded with the Lumen Prize 2022, the iF Design Talent Award 2022, the ADC Talent Award in Gold 2022 and the UX Design New Talent Award 2022.
Faculty of Design Würzburg
Bachelor Project by Max Seeger
First examiner: Prof. Erich Schöls
Second Examiner: Marcus Kaiser
Many thanks to Matthias Burba for his great support with all taxonomic questions within this project, as well as questions of microscopy and microphotography. A big thank you also to Dr Michael Kloster for insights into his research on AI-based diatom recognition and to Jan Rosenboom for the loan of his laid diatom specimens.
Bachelor Project by Max Seeger
First examiner: Prof. Erich Schöls
Second Examiner: Marcus Kaiser
Many thanks to Matthias Burba for his great support with all taxonomic questions within this project, as well as questions of microscopy and microphotography. A big thank you also to Dr Michael Kloster for insights into his research on AI-based diatom recognition and to Jan Rosenboom for the loan of his laid diatom specimens.